PORTFOLIO
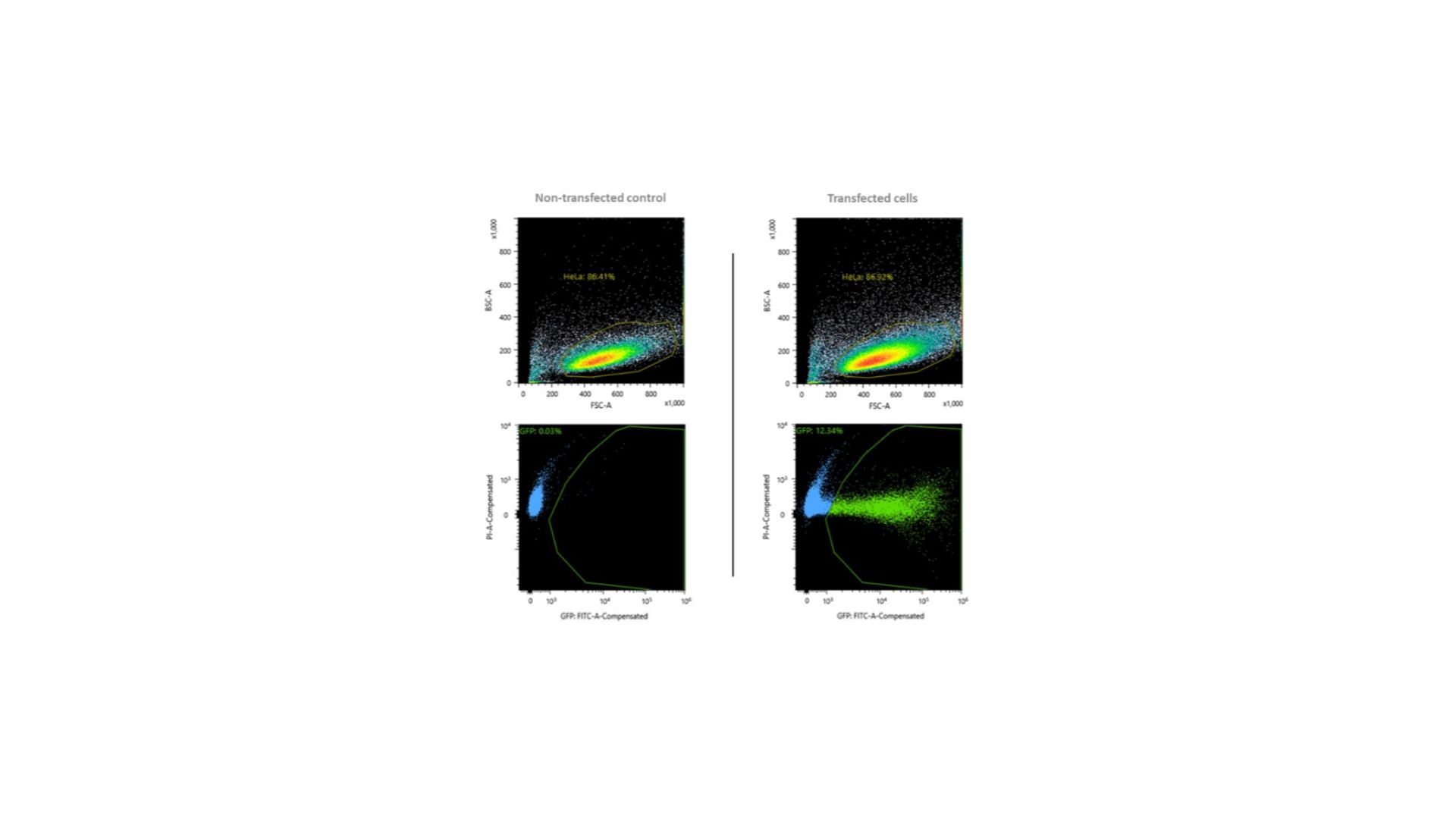
Selection of GFP+ cells 48h post-transfection with RNPs and GFP-encoding plasmid. 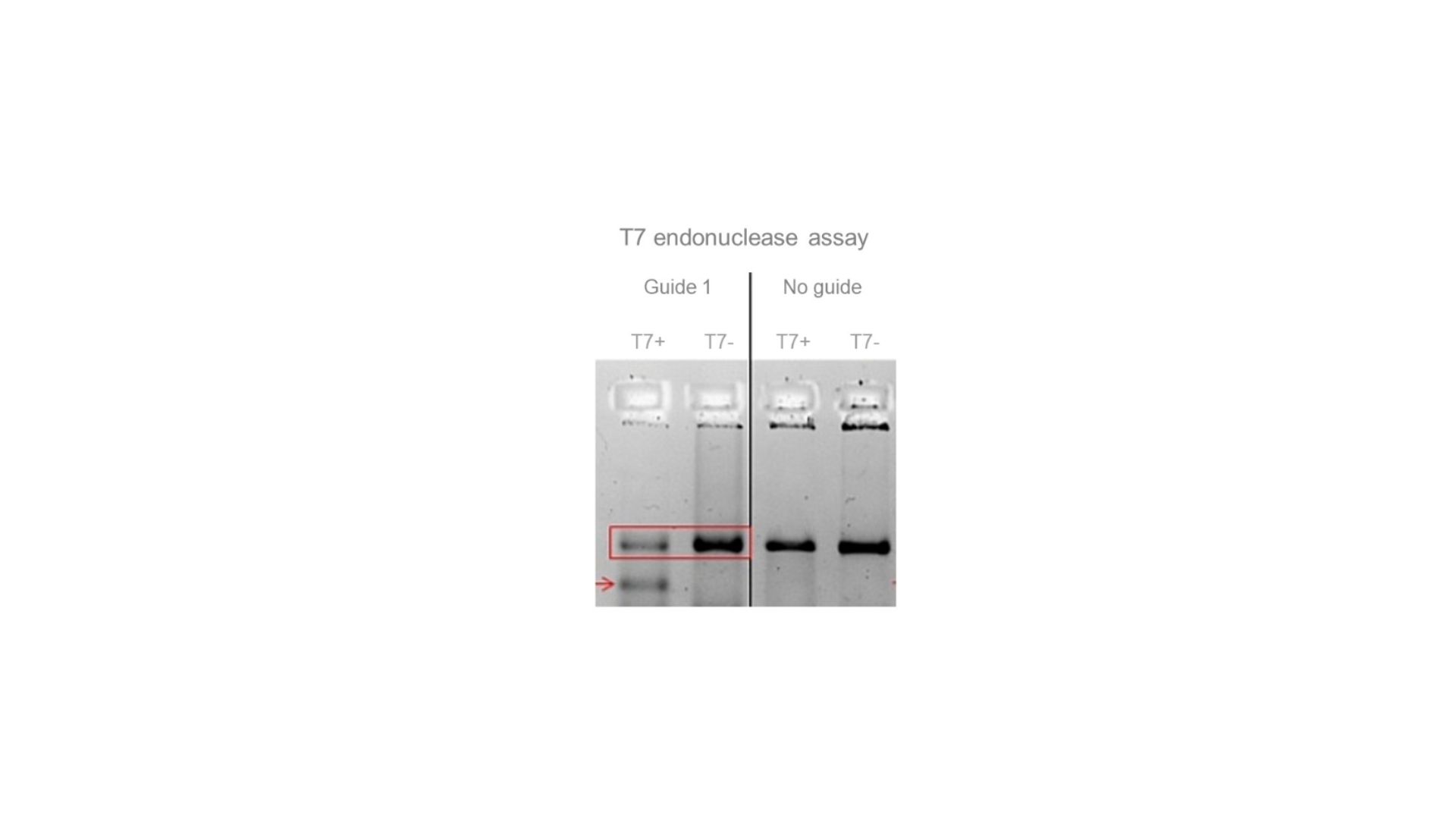
(Pre-)genotyping of a KO using T7 endonuclease (in bulk cultures and/or in clones). The red arrow indicates one of the bands that is produced by the endonuclease after digestion of a CRISPR-edited amplicon. 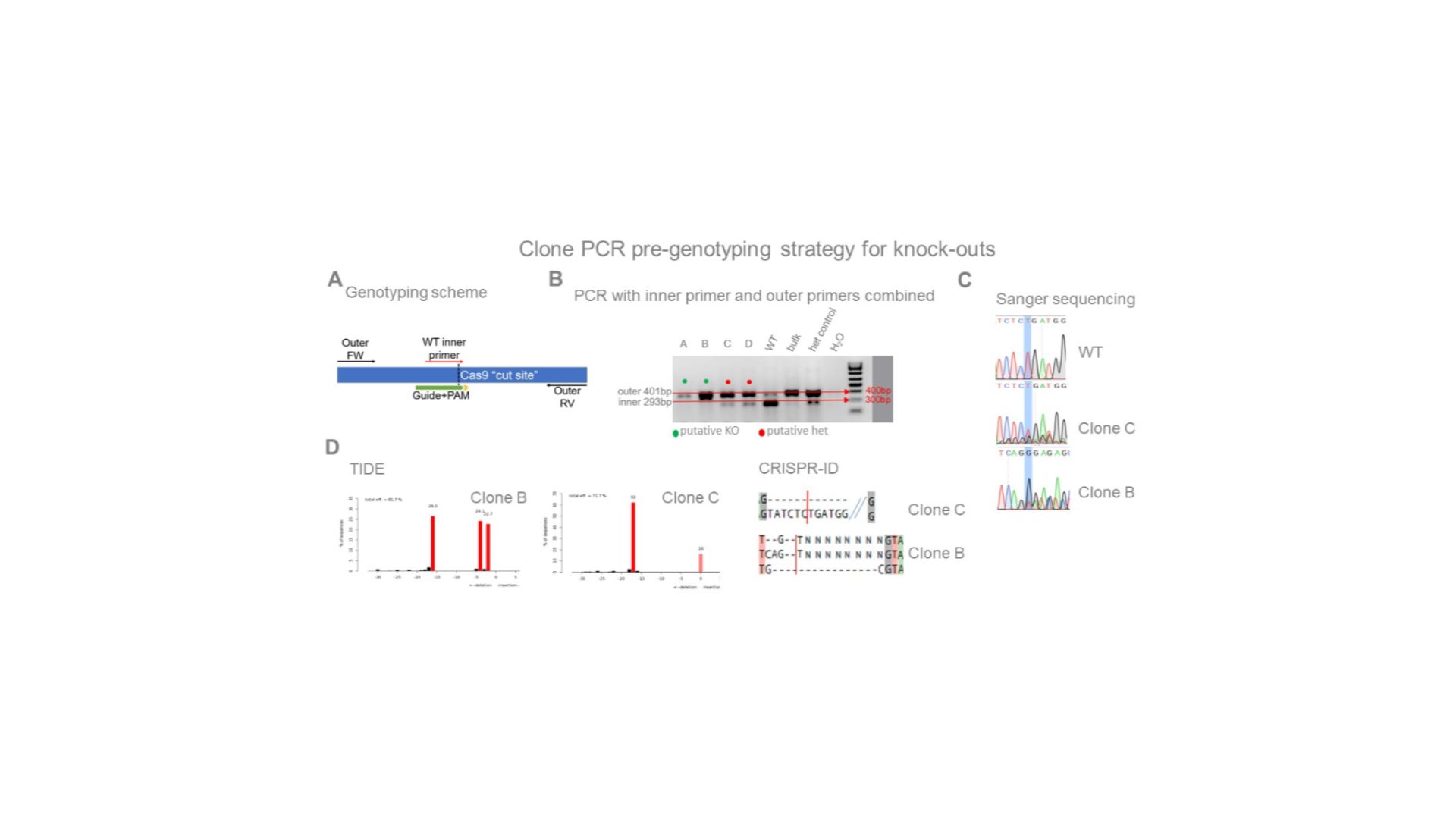
The WT inner primer (in A) overlaps the cut site in its 3′ end and will not produce the 293 bp amplicon when combined with the outer RV primer if all alleles have been cut and extensively edited. B) The lowest ratios of the 293 bp/401 bp amplicon intensities identify the most likely clones to have accumulated frameshifts in all alleles. C and D) This strategy has been validated by subsequent sequencing. As expected, clone B has all alleles edited and clone C still has one wild-type allele. 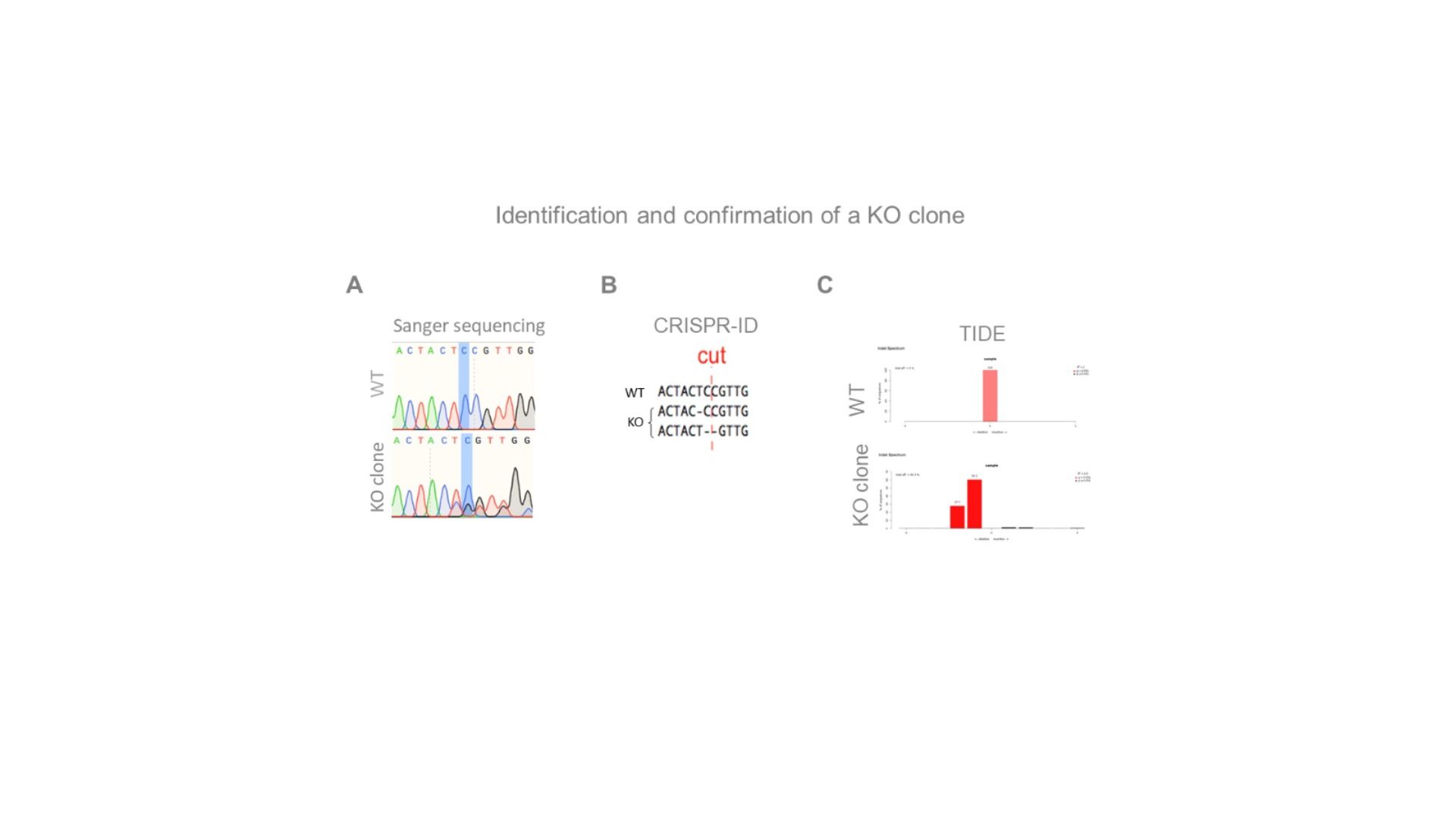
Highlighted nucleotide and “cut” indicate the expected Cas9 cut-site. CRISP-id and TIDE are programsthat identify the different allelic sequences present in the chromatogram. 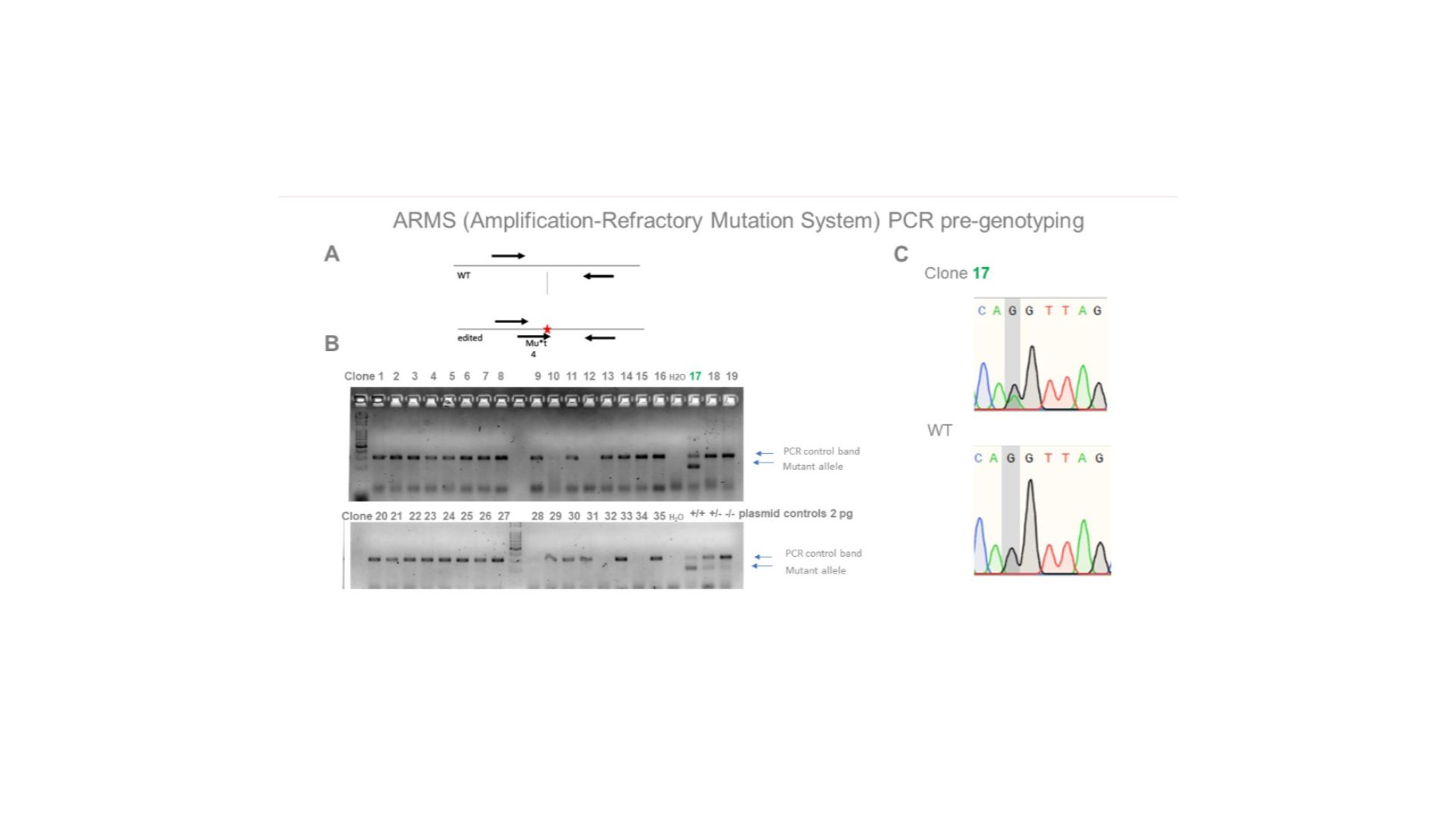
Single base substitution in an iPSC line. A) PCR genotyping scheme, with a primer specific for the altered allele. B) PCR data for clones. Clone 17 is a putative heterozygous clone. C) Confirmation by sequencing that the desired edit was introduced (in a heterozygous clone). 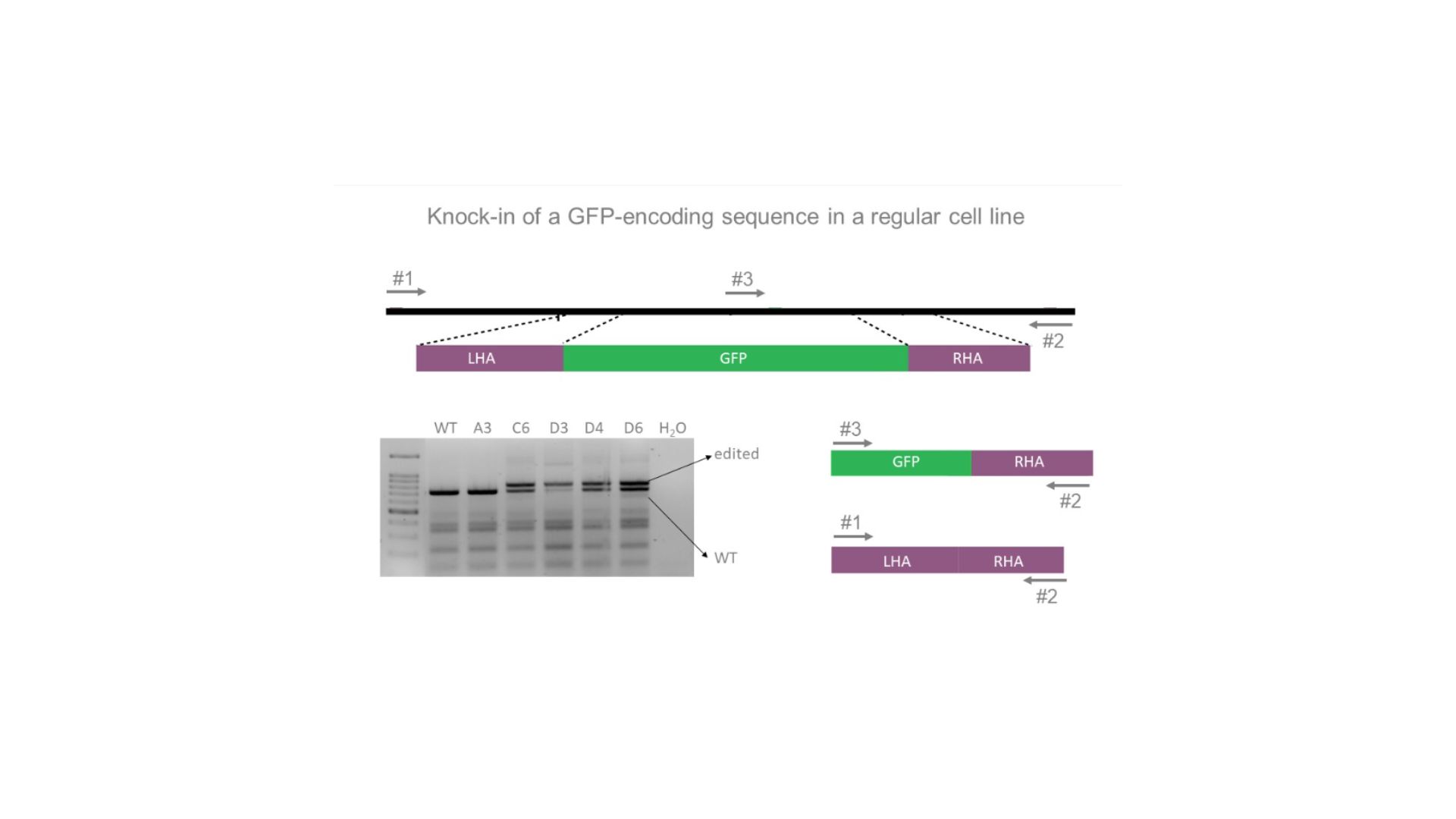
PCR genotyping of GFP gene insertion using two outer primers and one inner primer specific for GFP.LHA: left homology arm; RHA: right homology arm. 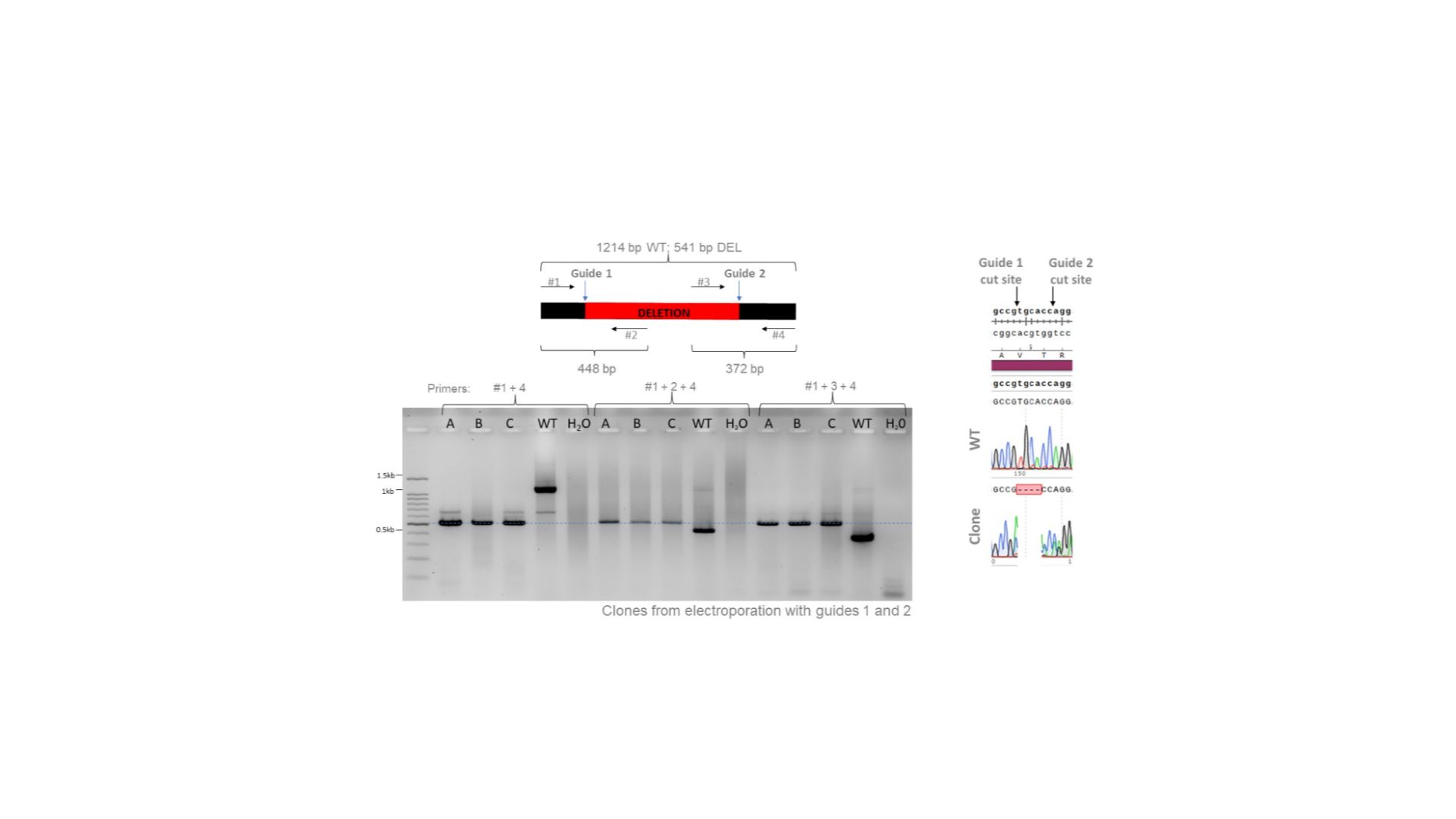
Clones from electroporation with guides 1 and 2 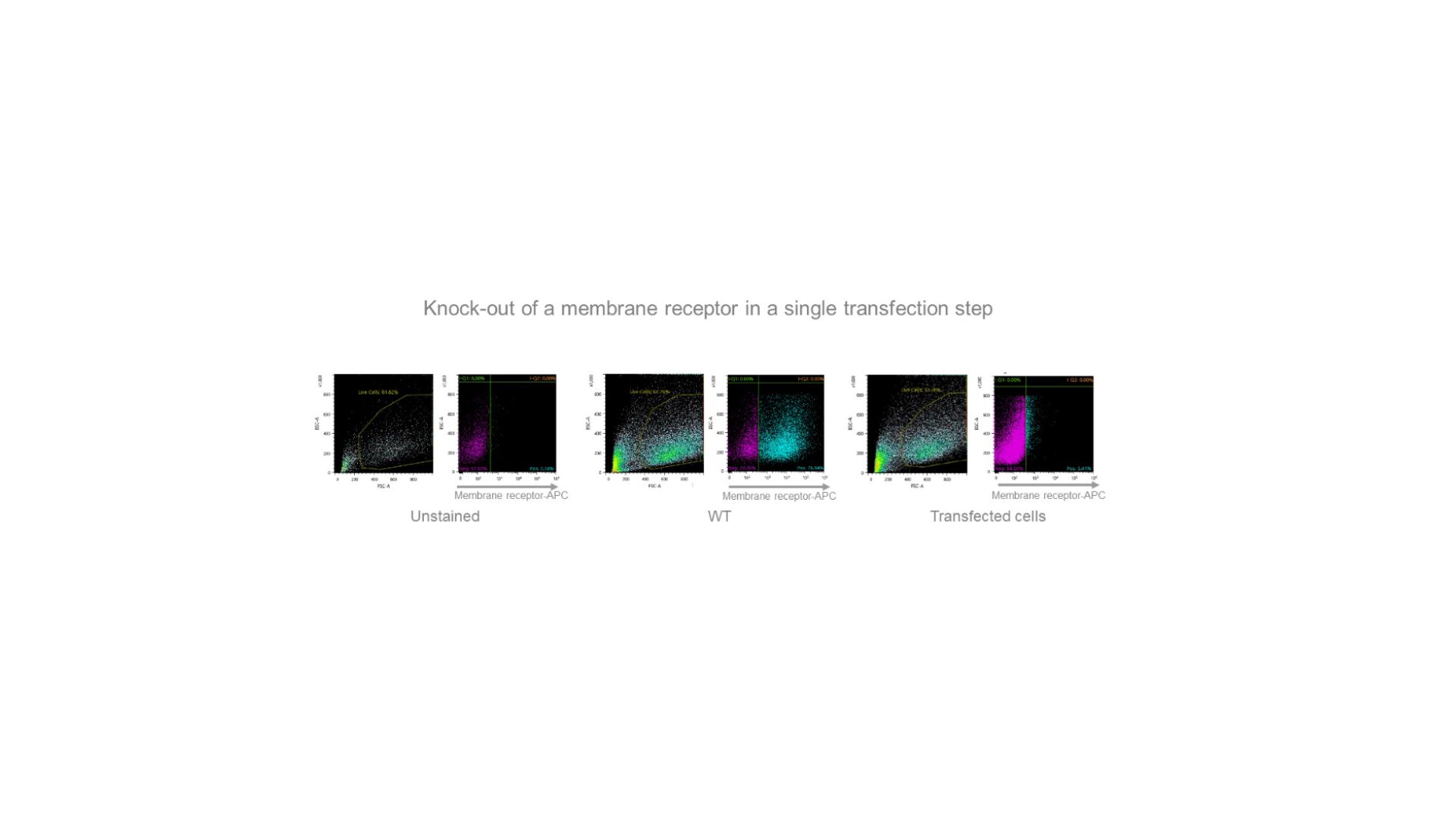
An exceptionally efficient sgRNA allowed us to produce a quasi-homogeneous population of cells “knocked-out” for a membrane receptor gene after electroporation of ribonucleoprotein particles (RNPs) and isolation of the transfected cells (APC antibody staining). 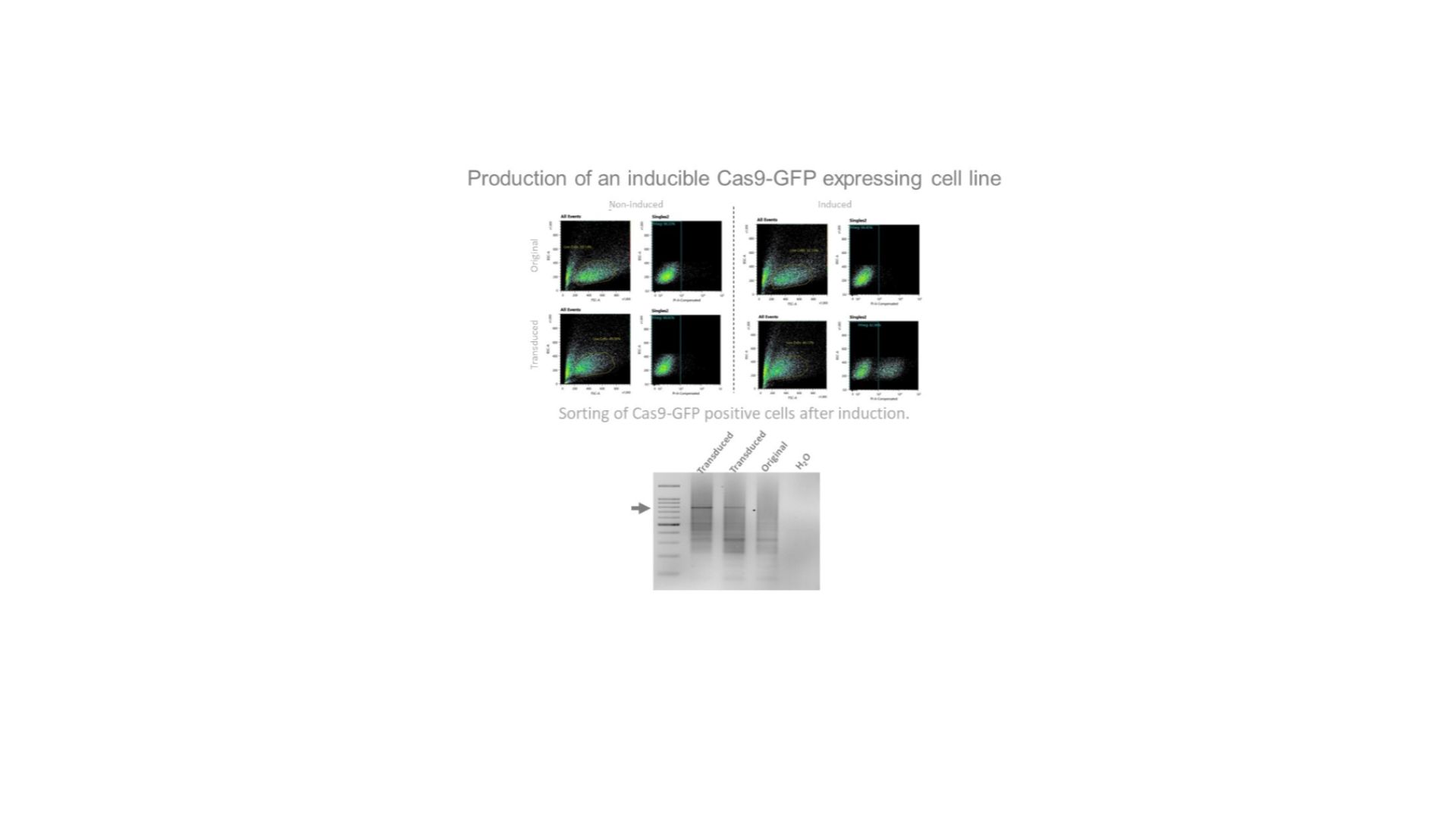
Confirmation of integration into the genome by Cas9-specific PCR.
